Future Products
|
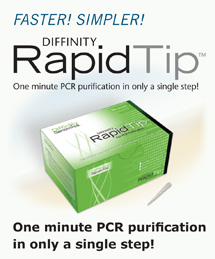
Sanger Sequencing Reaction Cleanup Are you a core facility seeking a faster, easier way to purify cycle sequencing reactions? Diffinity is developing functional pipette tips for the cleanup of cycle sequencing reactions (eg, BigDye cleanup) in just one minute! Purify a 96-well plate of BigDye reactions in less than two minutes using a compatible automated liquid handling robot. Use the purified samples directly in your capillary electrophoresis equipment. Interested in beta testing when it becomes available to test?
|